Testing of reliability of NevGen predictor
April 2017 testing, and general notes
During this month was performed the first testing of NevGen's prediction reliability, using SNP-analyzed samples from several projects, whose haplotypes are known to which subclade they belong. For testing were used only samples which were not known previously to NevGen, and which are not part of NevGen's statistics for it's subclades.
Not all such haplotypes were used, haplotypes which are too close to any haplotype that is part of statistics of it's already known subclade are ignored. For example, if we have 67 markers-long haplotype called H1 whose known subclade (supported by NevGen) is called SUBCL1, and in NevGen's statistics for SUBCL1 exists haplotype H2 which is less than 6 values distant from H1 (genetic distance is less than 6), than H1 is ignored and not used for testing. For haplotypes of 37 markers ignored are those with 3 or less such differences, and for those of 111 markers ignored are those with 8 or less. Since almost all such haplotypes are right predicted (although NevGen's prediction algorithm has nothing to do with genetic distances), this way percentages of right predicted haplotypes are lowered, and this results are rather conservative. Number of such haplotypes is not small, they were not counted, but in R1b U106 testing there was probably between 50 and 100 of them.
Also, haplotypes that belong to subclades which are not supported by NevGen (usually smaller ones, with too few SNP-proven haplotypes, with no or less than three full 111-marker haplotypes, or only with haplotypes of the same surname...) are ignored. Since their usage in predictor yields false positives.
Is need only to add when haplotype is considered "right" or "wrong" predicted by predictor in this testing. For simplicity, it is considered "right" if it's already known subclade has been given the greatest probability (and ofcourse if it is greater than zero), which means if it is the first in results list. In many cases, SNP-proven subclade were given substantiate probabilities (for example more than 20% or 30%), but nevertheless they are considered wrong if they are not the first in results list.
After testing was finished, all haplotypes used in testing are added to statistics of NevGen's supported subclades, whether they were wrong or right predicted. That way predictor's statistics is made better.
Carefull user might notice that performance of 111-marker haplotypes is, unexpectedly, not much better (or even worse) than performance of 67, or sometimes even 37 markers (case of R1a). From logical point of view, it is expected that more markers means more reliable predictions, and greater percentage of right predicted 111-markers haplotypes than those with 67. But, that would be the case only if NevGen's statistics holds only haplotypes with 111 markers, which is not case. In some haplogroups and subclades there are only smaller percentage of haplotypes with full 111 markers. Majority of haplotypes from poorer countries (noneuropean, Eastern Europe, Southern Europe and Balkans) has at most 67 markers. In some countries even 67 markers are not majority. And shortage of 111-marker haplotypes is reason why statistics for markers 68-111 is not so good as for those from 1 to 37. Because every 111-markers haplotype is also 67- or 37-markers haplotype, but vice-versa does not stand. There are many nonactive NevGen's subclades (now not used, because statistics for NevGen-supported subclade cannot be made without at least one full 111-markers haplotype) with status "waiting for any 111-markers haplotype to show up". In G haplogroup there is one subclade with more than 20 haplotypes in statistics, but with only one with 111 markers.
I1 from Sweden
First it was done with haplotypes of I1 from Sweden Project, which suited to strict rules defined above. There were 41 of them, plus additional six from Ulster Heritage Project, which makes total of 47. Haplotypes were from different length. 39 out of 47 are considered right predicted, which makes 82.98%.
Here is structure by length of those 39 who are considered "right predicted" in this testing:
1 x 12 markers
10 x 37
16 x 67
12 x 111
Among those considered right predicted is one with only 12 markers, with probability of 95.81%, which surprised us much. It belongs to subclade FGC10430.
Now structure of eight considered wrong predicted:
1 x 12 markers
3 x 37
2 x 67
2 x 111
Of 8 wrong, three were cases with very close subclades given highest probability. For example haplotype with 37 markers gave such results, when right subclade S14887> FGC21980 is on second place with 12.71%, and on the first place is very close subclade S14887> S18218:
I1 P109>Y3662> S14887> S18218 55.96%
I1 P109>Y3662> S14887> FGC21980 12.71%
Other 5 haplotypes gave result with more distant subclades on the first place. Wrongly predicted haplotype with 12 markers gave next results, whose confirmed subclade is L813:
I1 P109> Y3662>Y4045 31.03%
I1 L22>L813 15.45%
...
R1b U106
Now we go on results of testing with R1b U106 haplotypes from U106 Project. R1b is much harder nut to break. There are 168 haplotypes in total used in testing. 123 of them are considered right predicted, which makes 73.21%.Here is structure by length of those 123 who are considered "right predicted" in this testing:
1 x 12 markers
11 x 37
36 x 67
75 x 111
Structure of 45 considered wrong predicted:
15 x 37 markers
11 x 67
19 x 111
Here too many wrong predicted sample haplotypes gave most of their probability to very close subclades, like in next case where haplotype with 111 markers belongs to S1911:
R1b U106>Z381> Z156>DF98> S18823 55.64%
R1b U106>Z381> Z156>DF98> S1911 14.71%
In some cases biggest probability went to subclades which are not even U106, like in this case when 111-markers haplotype belongs to FGC8512:
R1b L51>L151> CTS4528> S14328 25.63%
R1b U106>Z381> Z301>FGC8512 12.82%
But, in majority of wrongly predicted cases, biggest probability went to subclades of U106. Nevertheless, results of U106 predictions are not satisfying in general. Many subclades in NevGen's statistics are represented with small number of samples, and their number must be increased in order better prediction to be achieved.
R1b samples from Ulster Heritage Project (mostly L21)
From publicly available haplotypes in Ulster Heritage Project used were 49 R1b samples, of whose 38 are considered right predicted, which makes 77.5%.Structure of 38 who are considered "right predicted" in this testing:
5 x 37 markers
13 x 67
20 x 111
Structure of 11 considered wrong predicted:
6 x 67 markers
5 x 111
Of those 11, nine were cases when SNP-proven subclade is downstream of M222, and the first subclade predicted is very close another subclade downstream of M222. Like in this example, where 67-markers haplotype belongs to S588.
R1b L21>DF13> Z39589>DF49>> M222>FGC4077 46.16%
R1b L21>> Z39589>DF49>> M222>S658>> S588 32.47%
Subclades of M222 are very young, and even more hard to distinquish among themselves, even on 111 markers. Nevertheless, NevGen recognized right subclades in more than half of samples (12) downstream of M222.
Ulster's R1b-s gave much better score than R1b U106 haplotypes, not only by percentage (77.5% vs 73.21%), but more significantly, in structure of those which are considered wrong predicted. Those from Ulster, which are mostly downstream of L21, in all 11 cases gave highest probability to very close subclades, which was not allways the case with haplotypes from U106. What is reason for such difference in quality of prediction? Most probably it is because average number of samples per subclade is considerably greater in subclades typical for British Isles (where L21 is very strong), than in subclades of U106, with more uniform spread across Northern Europe. So, it is clear, in order to be more reliable, predictor needs more samples for it's statistics! It is obvious when huge U198 subclade of U106 is considered. It has highest number of samples in statistics of all U106 subclades in NevGen, and till now, no sample SNP-tested positive for it (with at least 37 markers) was predicted wrong in NevGen, at least by us.
In some weeks similar testing of NevGen's R1b prediction shall be made on some even bigger public Project, like Irish, Scottish, or British Isles.
R1a from Sweden
For R1a prediction testing was used 36 haplotypes from Sweden Project, with two additional from Ulster Project (who are descendants of Scandinavians anyway), 38 in total. 35 of them are considered right predicted, or 92.1%.Structure of right predicted:
1x12
10x37
9x67
15x111
Structure of wrong predicted:
1x67
2x111
For all three wrong predicted sample haplotypes top probability went to very close subclades, like in this example:
R1a Y2395>Z284> L448>CTS4179 (all others) 61.24%
R1a Y2395>Z284> L448>CTS4179> YP386 16.97%
R1a Y2395>Z284> L448>CTS4179> YP704 1.67%
May 2017 testing of prediction of R1b from British Isles
In the first week of May 2017 NevGen R1b Level was tested using deep SNP-analyzed haplotypes from three public FTDNA projects of British Isles background, which were previously unknown to NevGen's statistics.
Projects used here are:
Ireland Y-DNA Project
Scottish Y-DNA Project
British Isles DNA Project by County
All haplotypes used in testing are known which subclade they belong of subclades supported by NevGen R1b Level. Rules of testing and criteria for selecting haplotypes are already described in General Notes in begining of this page and shall not be repeated here. 219 haplotypes in total passed strict rules, and 165 of them are considered right predicted, or 75.34%. Beside them other 168 haplotypes (which were all right predicted) were rejected because they were too close to some haplotypes from their already known subclade supported by NevGen (distance less than 4/37, 6/67 or 9/111).
Here is structure by length of those 165 who are considered "right predicted" in this testing:
20 x 37
48 x 67
97 x 111
Structure of 54 considered wrong predicted:
12 x 37 markers
23 x 67
19 x 111
Majority of tested haplotypes belong to L21 subclade of R1b, as is expected for persons of British Isles background. Substantial part of cases with wrong prediction goes to errors with distinquishing among subclades of M222. Final result (75.34%) of right predicted haplotypes is relevant only for haplotypes with origins from British Isles. For haplotypes of R1b from another parts of Europe such rate is not expected, but is expected to be less, due to less available haplotypes.
February 2018 testing of prediction of R1b L21
In the first week of February 2018 NevGen R1b Level was tested using deep SNP-analyzed haplotypes from L21 public FTDNA project, which were previously unknown to NevGen's statistics.
Like in earlier testings, all haplotypes used in testing are known which subclade they belong of subclades supported by NevGen R1b Level. Rules of testing and criteria for selecting haplotypes are unchanged, they are already described in General Notes in begining of this page. In total 71 haplotype passed strict rules, and 54 of them are considered right predicted, or 76.06%. Beside them other 49 haplotypes (which were all right predicted) were rejected because they were too close to some haplotypes from their already known subclade supported by NevGen (distance less than 4/37, 6/67 or 9/111).
Here is structure by length of those 54 who are considered "right predicted" in this testing:
2 x 37
4 x 67
48 x 111
Structure of 17 considered wrong predicted:
1 x 67
16 x 111
Here we can see that percentages of "right predicted" haplotypes are alike another testings of R1b Predictor. All 17 haplotypes considered "wrong predicted" gave the greatest probability to another L21 subclade. In 5 out of 17 cases, right subclade was on second place, like in this case.
Probability = 51.64% Fitness=66.70 [0.92] R1b L21>FGC11134>> S1121>> Z18170
Probability = 32.27% Fitness=66.34 [0.99] R1b L21>FGC11134>> A151
Among "right predicted" are two haplotypes with 111 markers which belong to S1051 subclade. They were very, very distant from another haplotypes in NevGen's statistics, 29 and 36 markers from it's closest. Such thing happpens seldom in R1b. In another haplogroups sometimes happens that NevGen right predicts haplotypes which 40+/111 or even 50+/111 markers distant from it's nearest in predictor's statistics, but such distances are hard to find in (western) R1b. This is result of that with distance of 29:
Probability = 52.71% Fitness=46.49 [0.82] R1b L21>DF13> Z39589>S1051
Probability = 0.00% Fitness=41.96 [0.77] R1b L21>DF13> FGC5494
From our experience, it seems that rates of "right predicted" haplotypes of U106, DF19 and DF99 are similar as those of L21. Rates in Z2103 might be even greater. But, rates in DF27 and U152 are much lower, especially in former, where I think they go to less than 30%. Those two are so unreliable partially due to less available deep-SNP tested haplotypes. But, even in them there are some subclades which are much more righly predicted. Unsurprisingly, those are subclades which are common in British Isles.
2019 testing of prediction, July 31st 2019
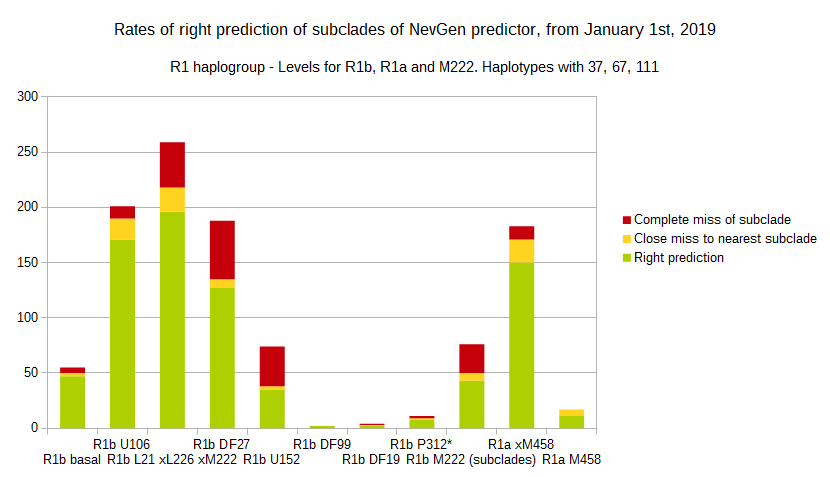
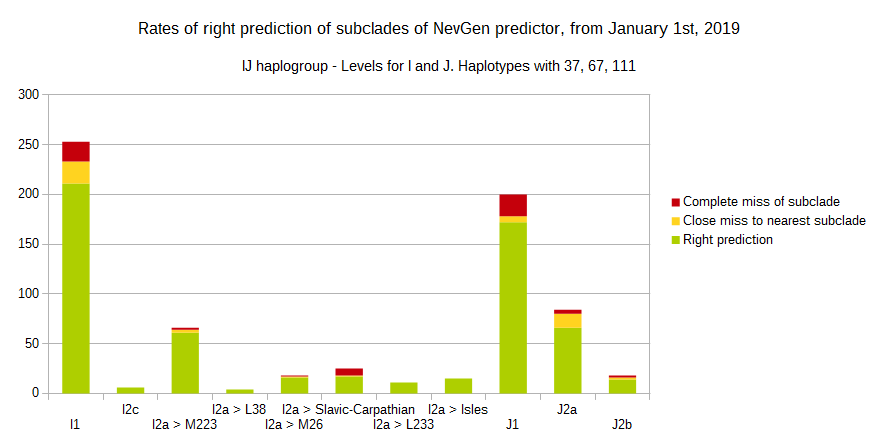
Since January 1st 2019, we have started to record how newly found haplotypes with known deep SNPs are predicted for (their already known) subclade with NevGen Predictor. Statistics of it you can see on the next three pictures.
First is for haplogroups R1b and R1a, second is for I and J, and third is for all others.
To avoid confusion, should be noted that statistics is concerned with prediction of deeper subclades of mentioned higher-level haplogroups, not for prediction of higher-level haplogroups themselves. For example, I1's 83% of right subclade predictions is for prediction of subclades of I1, not for prediction of I1 alone, which is easy with at least 25 markers. The same holds for all other haplogroups from pictures, like I2a > L38, I2a > Isles, I2a > M223, J1, E > V13, R1b > M222, R1a M458, C, L, Q, T, D and so on.
Important thing is that all haplotypes which are less than 9/111, 6/67 or 4/37 close to any haplotype which is part of statistics of it's already known subclade are excluded from this statistics.
That way statistics is more reliable, since such close haplotypes are almost always good predicted. Haplotypes with less than 37 markers were not used in this testing, but majority of them had 111 markers.
Level for R1b-M222 used here is not available in public, since we are not satisfied with it's results.
From statistics for R1b-L21 are excluded all haplotypes of M222 and L226, because they are trivial to predict themselves (but not their deeper subclades).
As we can see, the worst prediction results are for R1b-U152. I believe we need many hundreds (if not thousands) of new haplotypes to get better results.
With yellow colour we have marked haplotypes which got the most of probability to subclade which is very close, one level below. For example, if our haplotype is proven to belong to "R1b U106>Z381> Z301>L48> Z9>>Z326>> A5011", but we got top probability in NevGen for "R1b U106>Z381> Z301>L48> Z9>>Z326>> BY4305", we than record this testing as "close miss".
From our experience, subclades under L1335 and FGC11134 (R1b > L21), and are not easy to be distinguished, and also Z326 under U106 or L1029 (R1a > M458) or L258 (I1), just to mention some of them.

2020 - 2021 testing of prediction, December 13th 2021
We have prepared new results of testing of NevGen Predictor. Time span is from June 21st 2020 to July 31st 2021. Little more than 13 months.
As was done in previous years, newly found haplotypes with known deep SNPs are predicted for (their already known) subclade with NevGen Predictor. Statistics of results can be seen on the next eight pictures (four are by number of haplotypes used, and other four by percentages). Haplotypes which do not belong to any of supported subclades (due to small number of available samples) are not used in this statistics. As always, prediction was considered as right if haplotype belongs to subclade which is the first in list (it's already known subclade has greatest probability), and if it's probability is greater than 0 (please not that probability do not need to be 100%, but must be the first and must be bigger than 0).
First is for whole of haplogroup R (R1b, R1a and R2), second is for haplogroups I and J, third is for haplogroups E and G and fourth is for N and all other haplogroups. As we can see, percentages of right predictions are similar to prediction rates of previous year.
Like we said before, to avoid confusion, should be noted that statistics is concerned with prediction of deeper subclades (those that NevGen Predictor supports) of mentioned higher-level haplogroups, not for prediction of higher-level haplogroups themselves. For example, I1’s 75.8% of right subclade predictions is for prediction of subclades of I1, not for prediction of I1 alone, which is easy with at least 25 markers. The same holds for all other haplogroups from pictures, like I2a > L38, I2a > Slavic-Carpathian, I2a > Isles, I2a > M223, J1, E > V13, E > M84, N > VL62, R1b > M222, R1a M458, C, L, Q, T, D and so on.
Important thing is that all haplotypes which are less than 9/111, 6/67 or 4/37 close to any haplotype which is part of statistics of it’s already known subclade are excluded from this statistics. That way statistics is more reliable, since such close haplotypes are almost always good predicted. Haplotypes with less than 37 markers were not used in this testing, and majority of them had 111 markers.
Level for R1b-M222 used here is not available in public, since we are not satisfied with it’s results. From statistics for R1b-L21 are excluded all haplotypes of M222 and L226, because they are trivial to predict themselves (but not their deeper subclades).
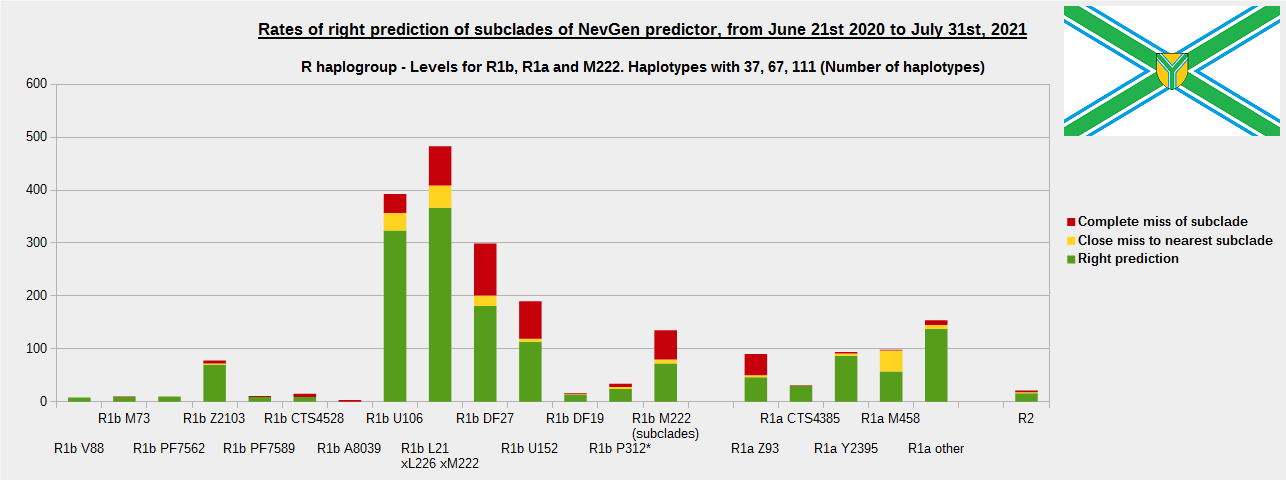
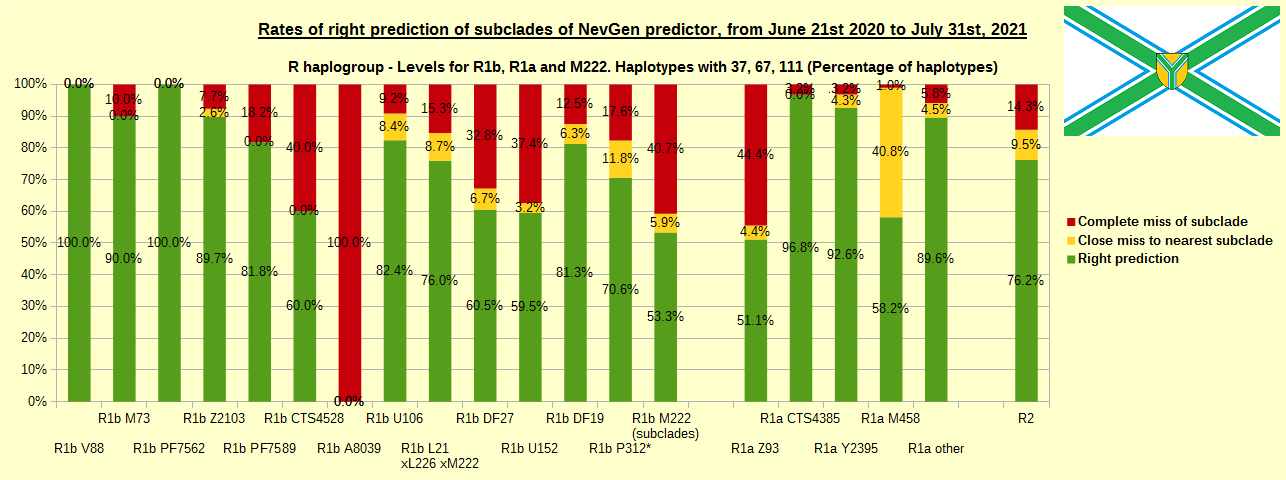
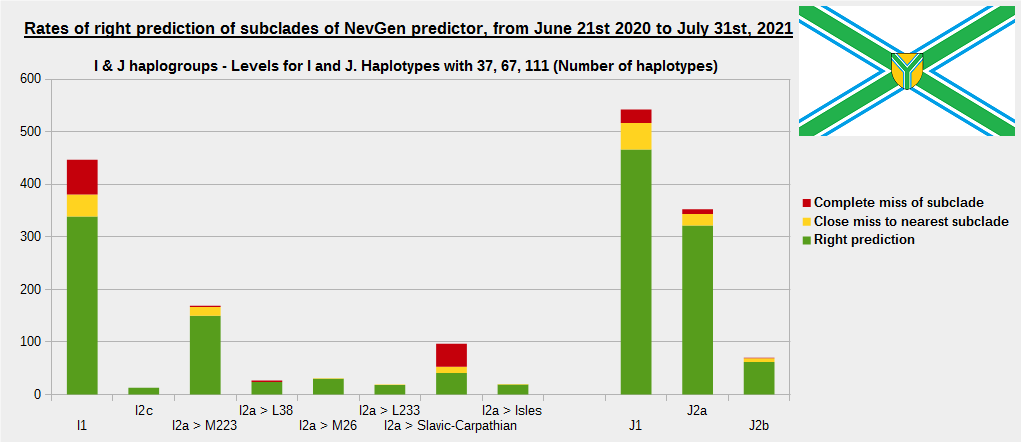
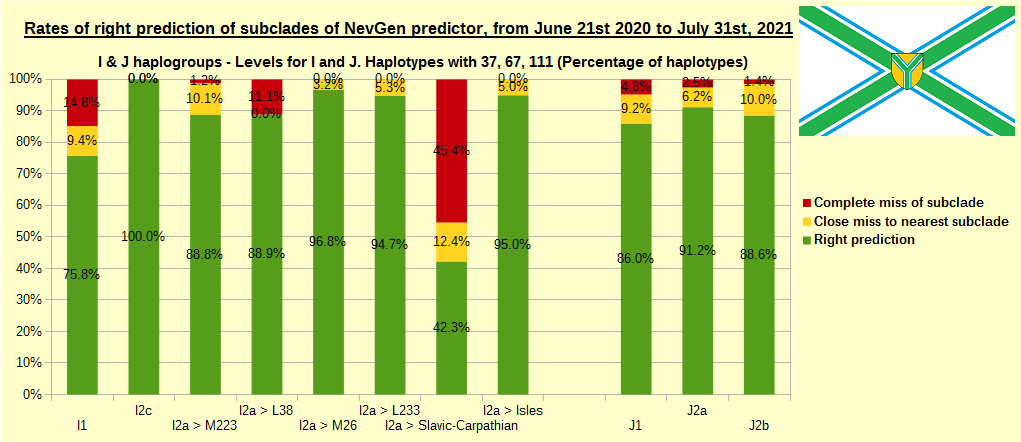
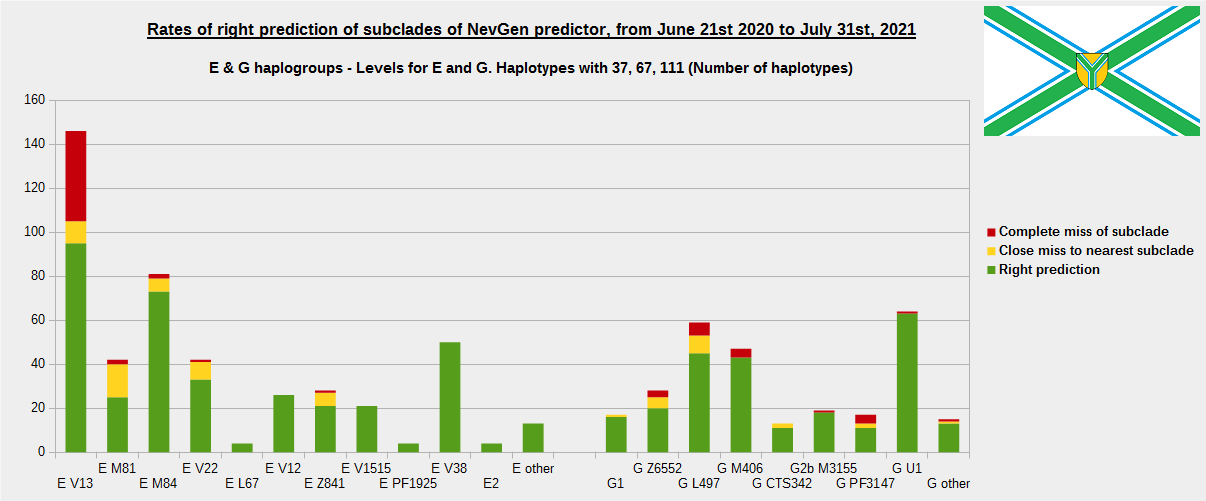
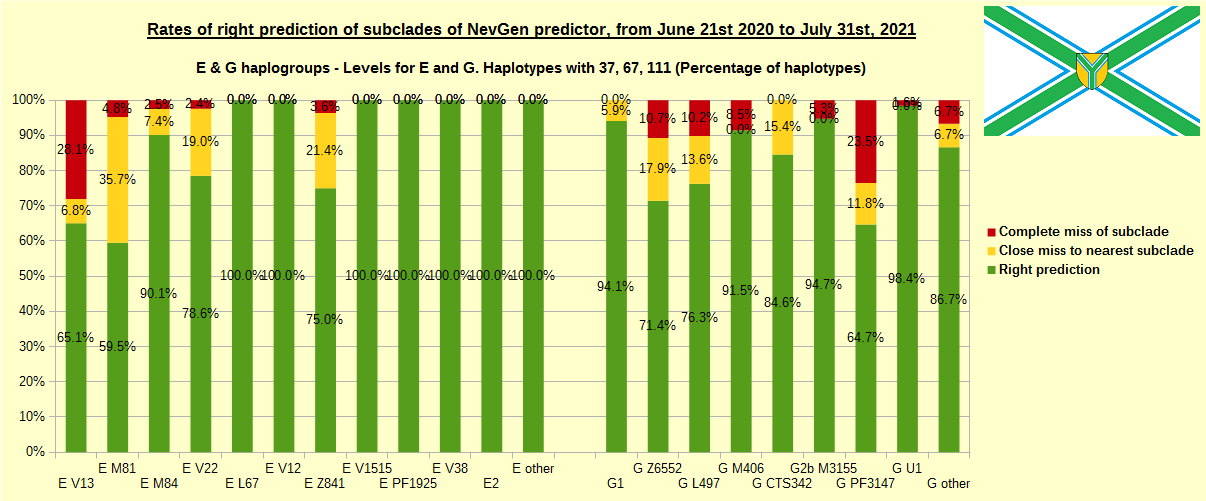
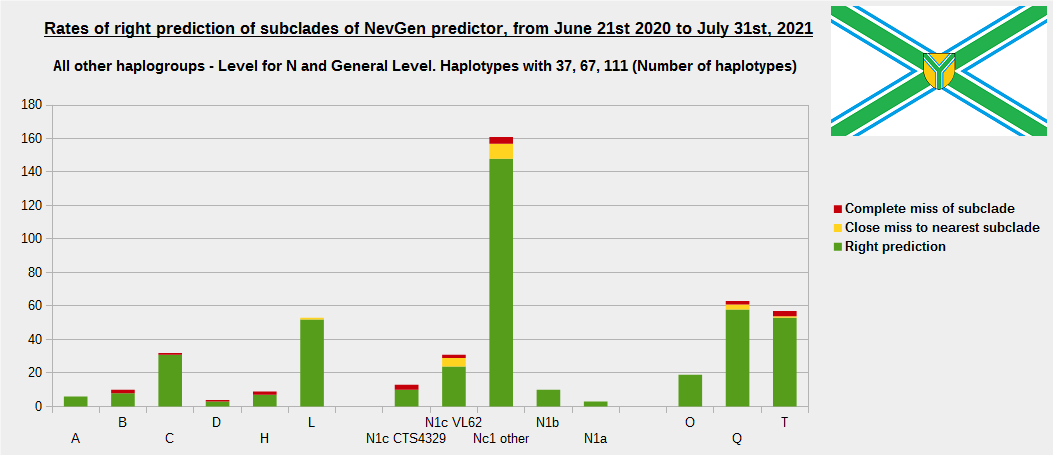

In this year, the worst prediction rates of major haplogroups are for I2a > Slavic-Carpathian (my own hg, long time ago wise men said that man cannot be prophet (in our case "predictor") in his own village). Here now we have more wrong predicted than right predicted, last year's results for this haplogroup were slightly better. This year second place as the worst prediction results are for R1b-U152, 59.5% (last year it was the worst), similar to last year's 59.7%. I still believe we need many hundreds (if not thousands) of new haplotypes to get satisfying results for U152.
Like in the previous years, with yellow colour we have marked haplotypes which got the most of probability to subclade which is very close, one level below. For example, if our haplotype is by SNP proven to belong to “R1b U106>Z381> Z301>L48> Z9>>Z326>> A5011”, but we got top probability in NevGen for “R1b U106>Z381> Z301>L48> Z9>>Z326>> BY4305”, we than record this testing as “close miss”.
From our experience, subclades under L1335 and FGC11134 (R1b > L21), are not easy to be distinguished, and also Z326 under U106 or L1029 (R1a > M458) or under L258 (I1), just to mention some of them.
Now something we wrote first time last year: long-distance right predictions. That are predictions when haplotypes were right predicted, despite being very far from any sample in our statistics of it’s subclade. In our 13-months time-span now we had 393 of them (in last year it was 98), when distance from nearest is at least 30/111. Such cases mostly happened in haplogroups E and J2a, just like in last year.
Now our most distant successful prediction (and best of all times) is for 111-markers haplotype of haplogroup C1a2 V20, which is right predicted despite being 96 markers far from its nearest in training dataset for its haplogroup (it was predicted with only 1.12% probability, but no other clade got more than 0).
Second most distant successful prediction is now in haplogroup E1a M132, where distance to nearest was 84 (2.96%), and third is in haplogroup C1 F3393, where distance to nearest was 83 (1.99%). Of course,in all cases this was the greatest probability given of all clades. In R1b, the greatest distance with right prediction was 48, and haplotype belonged to subclade R1b V88 >> FGC21039.
In previous year two biggest distances were 78 and 71 (in haplogroups G2b M3155 and J2a > Z6048).
Here you can see whole list of such long-distance right predictions, by haplogroup, with distance from nearest, and in many cases it’s subclade.
A (4 haplotypes): 41, 57 (A1b1b2b M13), 37, 69 (A1a M31)
B (3 haplotypes): 32, 32, 59 (B2a1 M218)
C (10 haplotypes): 73, 76, 96 (C1a2 V20), 39, 45 (C1 > PH407), 83 (C1 F3393), 35, 35 (C2b1a1b1 F3985), 39, 50 (C2 M217> F1067)
D (2 haplotypes): (D1a1a1 PH4), 63 (D1b2a CTS220)
E (71 haplotypes). Distances: 30/111 (V13), 35, 44 (E1b1b V68> SK863), 36, 40 (E1b1b V1515), 34, 38, 39 (E1b1b >> M84>> Y5435), 31, 35, 35 (E1b1b V1515> V1700), 31, 36, 39, 39, 40, 40, 40, 41, 41, 41, 49 (E1b1a V38>> L485), 33 (E1b1a V38>> M58), 35, 36, 38, 42 (E1b1a V38>> M4231), 31, 31, 36, 36, 38, 40, 43, 48 (E1b1b M123>M34> M84>> PF6751> PF6748), 33 (E1b1b V22), 39 (E1b1b V22>> BY7640), 44 (E1b >> M281), 31 (E1b1b V13>>S7461), 33, 36, 48, 48 (E1b1b V22>> PH2818> BY7402), 54 (E1b1b V257> PF2431), 31 (E1b1b M123>M34> M84>> PF6751> Y6200), 42 (E1b1b M123>M34> Z841>> Y4971> Y4972), 31, 33, 34 (E1b1b V1515> CTS2871), 41, 44 (E1b1b V22>> PH2818> PH2121), 30, 33, 39, 39 (E1b1b V12>CTS693), 39, 40, 40, 40 (E1b1b V22>> PH2818> BY1984), 31, 32, 38 (E1b1b V12>V32), 31 (E1b1b V13 >> Y19509), 32 (E1b1b V13>>Z5018> L17), 38 (E1b1b L618>BY28614), 31, 54 (E1b1b V1515> PH1818), 30 (E1b1b V13>>Z5018> S2979> Z16659>Y3183), 41 (E1b1b M123>M34> M84), 44 (E1b1b M123>M34> M84>> Y5435), 84 (E1a M132)
G (52 haplotypes): 32 (G2b M3155), 37 (G2a2 > L497>> Z725>> Z16775> Z16777), 30, 34, 36, 37, 38 (G2a2b1 M406> PF3293), 45, 50, 50, 54, 60, 61, 62 (G1a CTS11562), 57 (G2a2a PF3147>> FGC6669), 41 (G2a2b2a1c CTS342), 34 (G2a2b2a1c CTS342> Z724>> Z26414), 67 (G2a2a PF3147> Z36520), 32 (L497>> Z725>> CTS4803> S2808> FGC8303), 30 (G2a2 > L497>> Z725>> L43), 31, 34, 37 (G2a2 > U1> L1266), 38, 42, 44, 45, 46 (G2a2 >> Z30503), 31, 32, 40, 41, 42, 42, 47 (G2a2b1 M406> FGC5081> Z17887), 34, 38, 48, 49 (G2a2b1 M406> M3302), 30, 34, 34, 36, 49 (G2a >> PF3359), 31 (G2a2 > U1> L13), 40, 42 (G2a2a PF3147>> L91), 30 (G2a1 Z6552 >> Z31464), 30, 32 (G2a2 > L497>> S10458), 38 (G2a2b1 M406> FGC5081> S9350), 32 (G2a2 > U1> L1266>> S9409), 52 (G2a1 Z6552 > L293 >> FGC1160)
H (2 haplotypes): 34, 64 (H1a1a M82)
I1 (2 haplotypes): 35 (I1 Z58>Z138>S2293>Z2541> Y7043), 30 (I1 Z58>Z138>S2293>Z2541> S19185)
I2 (17 haplotypes): 31, 33, 47 (I2a2a M223>Y4450>> M284>>Y3709), 32 (I2a2b L38> S2606> PH1237), 31 (I2a2a M223 >> S2364>Y4955), 31 (I2a1a Sardinian M26>F1295), 33, 40 (I2a1a Sardinian M26>PF4088), 32 (I2a2a M223>L701> P78> S25733), 31, 32 (I2c2 Y16419), 44 (I2a1 S21825> L1294 ("France")), 34 (I2a1a Sardinian M26>Y11222), 31 (I2a2a M223>L701> P78> S25733), 38 (I2a2a M223>Y4450> L1229> S18331), 33 (I2a2a M223>L701> L699> S12195), 36 (I2a2a M223>Y4450> L1229)
J1 (30 haplotypes): 43 (J1a >> P58>> S4924 >> L818), 31 (J1a >> P58>> L858>> Z640> BY74), 51, 55 (J1a >> PH77), 30, 31, 32 (J1a >> P58>> L860), 39 (J1a3 Z1828> Z1842> ZS3089), 32 (J1a >> P58>> Z18297>> Z18292),
34 (J1b F4306>> F1614>> F3547), 35 (J1a >> P58>> FGC11> FGC3723), 31, 34 (J1a >> P58>> S4924> ZS5379), 32, 33, 34, 34, 35, 38, 38, 42, 44 (J1a3 Z1828> Z1842> Z18436> CTS1460> CTS7188), 40 (J2a1 Z6063>> Z27802), 32 (J1a3 Z1828> Z1842> Z18436> ZS3042), 42 (J1a >> L620>> FGC6064>> BY97605), 53 (J1b F4306>> F1614>> Z2223), 32 (J1a3 Z1828> BY69), 43 (J2a1 Z6063>> Z27802), 32, 45 (J1a >> P58>> ZS1282)
J2a (69 haplotypes): 32, 34 (J2a1 M67>> S25258> S21160), 30 (J2a1 Z387> FGC35503), 26/37, 34, 39, 47 (J2a1 Z7700> FGC9883), 35, 41 (J2a1 PF5087> Z7430), 39, 40, 44, 46, 53, 53 (J2a2 PF5008 >> PF5040), 35, 65 (J2a1 Z6046> Z6048), 32 (J2a1 L26>Z500> M92> CTS4132), 38 (J2b1 M205 >> CTS1969), 38 (J2a1 M67>> S25258> SK1336), 31, 33, 33 (J2a1 Z387>L70> Z2177), 40 (J2a1 PF7431> L243), 33, 36, 36, 38, 40, 43, 48 (J2a1 Z7671> CTS900> CTS6804), 35, 39, 39, 42 (J2a1 Z7671> CTS900> Y11202), 30, 32, 38 (J2a1 Z7671), 33, 35, 38, 40, 41, 41 (J2a1 L26>Z500> M92> PF7412), 34, 37 (J2a1 Z6065> Y7708>M47), 30, 31 (J2a1 Z467> Z455), 35, 41 (J2a1 M319> Y92462), 32, 35, 37, 37, 39 (J2a1 Z6046> Y12610), 44 (J1a2 > ZS6591), 59 (J2a1 Z6065> Y7708> Z39478), 48, 51, 54 (J2a1 PF5191>> PF5177), 32 (J2a1 Z6063>> BY759), 34 (J2a1 Z387>L70> CTS3601), 30 (J2a1 Z6063>> BY759), 30 (J2a1 Z6063>> Y28259), 35 (J2a1 M67>> S25258> PH245), 31 (J2a1 PF5191>> S19231), 48 (J2a1 L26>Z500), 45, 62 (J2a1 Z6065)
J2b (5 haplotypes): 32, 34 (J2b2a M241>> Z1296> Z1297), 39 (J2b2b PH1648), 40 (J2b2a M241), 34 (J2b2a M241>Z2444)
L (13 haplotypes): 30, 32, 32 (L1a M27), 43 (L1b M317), 31, 38, 39, 39, 40 (L1c M357), 39 (L1b M317> M349), 18/37 (L1b M317> SK1412> PH1060), 39, 41 (L1b M317> SK1412> SK1414)
N (3 haplotypes): 31 (N2 P189.2), 35 (N1a2 CTS6380), 30 (N1a1 M46 >> M2019)
O (11 haplotypes): 33, 37, 38, 25/37, 46 (O2a2 F525), 35, 38 (O1b1 F2320), 60, 63, 28/67 (O2a1 F51), 45 (O1a1 B384)
Q (19 haplotypes): 34, 45, 52 (Q M346>> Y4800> F835> L932), 37, 37, 42 (Q F1096> M120), 46, 54, 61 (Q M346>> M3> M902), 39 (Q F1096> M25> F4531), 38 (Q M346>> L330), 37, 40, 42 (Q L275>> BZ50023), 42 (Q L275>> Y2265), 33 (Q L275>> L245> YP745), 39 (Q M346>> Y4800> Z5902), 39/67 (Q M346>> M3> Y4276), 33 (Q M346> YP4004)
R1a (7 haplotypes): 31 (R1a Z282>Z280> CTS1211>YP1034), 32 (R1a Z282> Y17491), 33 (R1a Z93>Z94>S23592), 36, 39, 46 (R1a YP4141), 33 (R1a Z93>Z94>Z2124>YP413)
R1b (59 haplotypes): 32, 34, 38, 42, 42 (R1b V88 >> V1589), 40 (R1b L21>FGC11134>> S1121>> Z18170), 30, 38 (R1b U152>L2> Z49>Z142> Z51), 33, 35, 40, 42 (R1b Z2103> Z2106), 34 (R1b L21>DF13> Z39589>S1051), 31 (R1b Z2103>Z2106>> PH4902), 34 (R1b Z2103>Y4362), 32, 35, 36 (R1b PF7562), 44, 48 (R1b V88 >> FGC21039), 32 (R1b Z2103>Z2106>> Y5587), 30, 35, 38 (R1b L51>PF7589), 32 (R1b U106>Z381> Z156>DF96), 31 (R1b L21>> ZZ10>Z253> Z2186>L1066), 34 (R1b Z2103>Y4362>> BY13762), 34 (R1b DF27>Z196> L176.2>L165), 30 (R1b DF27>ZZ12> Z2552> DF81), 32 (R1b DF27>ZZ12> ZZ19>Z31644> A2146), 33 (R1b L51>PF7589 >> CTS11824), 31 (R1b U106>Z381> Z301>L48> CTS3104), 31 (R1b U106>Z381> Z156>S5520> S5556), 31, 32, 33, 34, 35, 41 (R1b Z2103>L584> PF7580), 31, 32 (R1b P312>DF99> FGC847), 30 (R1b L21>> Z39589>DF49> ZP20> ZP80), 34 (R1b DF27>ZZ12> Z225> Z29884),
31 (R1b U152>L2> Z258>L20> Z1910), 33 (R1b Z2103>Z2106>> BY250), 34 (R1b M73 >> BY40441), 31 (R1b DF27>ZZ12> Z225> Y16019), 34, 35 (R1b Z2103>Y4362>> BY41455), 30 (R1b U152> L2> BY31138), 30 (R1b U152>L2> DF103> FGC8158), 31, 33 (R1b L21>> DF21>>DF5> ZZ32> CTS3655), 38 (R1b L51>PF7589 >> FGC24138), 30, 32 (R1b Z2103>Z2106>> BY250), 39 (R1b DF27>ZZ12> ZZ51> FGC49020), 37 (R1b L21>DF13> Z39589>Z251>> S11556), 35 (R1b L21>> Z39589>DF49> ZP20> ZP21)
R2 (7 haplotypes): 33, 34, 42, 29/67, 44, 47, 52 (R2 M479)
T (6 haplotypes): 38, 41 (T L131 > CTS933), 32, 44 (T >> CTS11451> Y4119), 40 (T>PF5633> Y16897), 48 (T >> CTS11451> Y4119>> Y3781)
2021 - 2022 testing of prediction, July 29th 2022
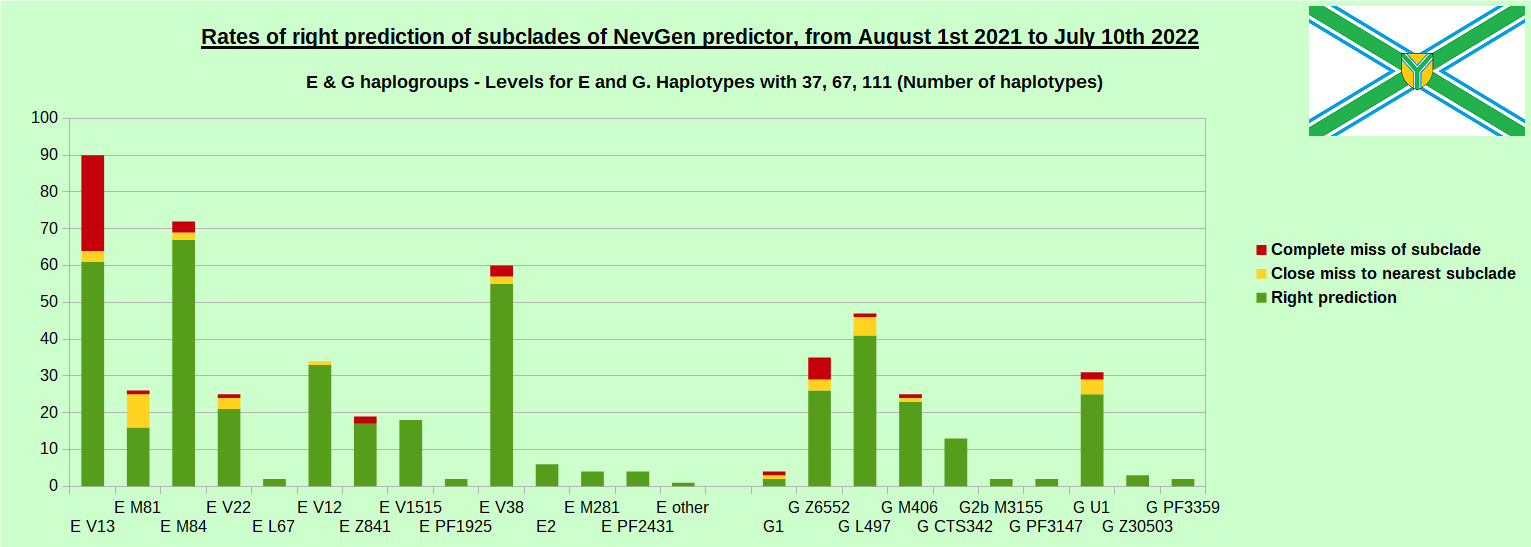
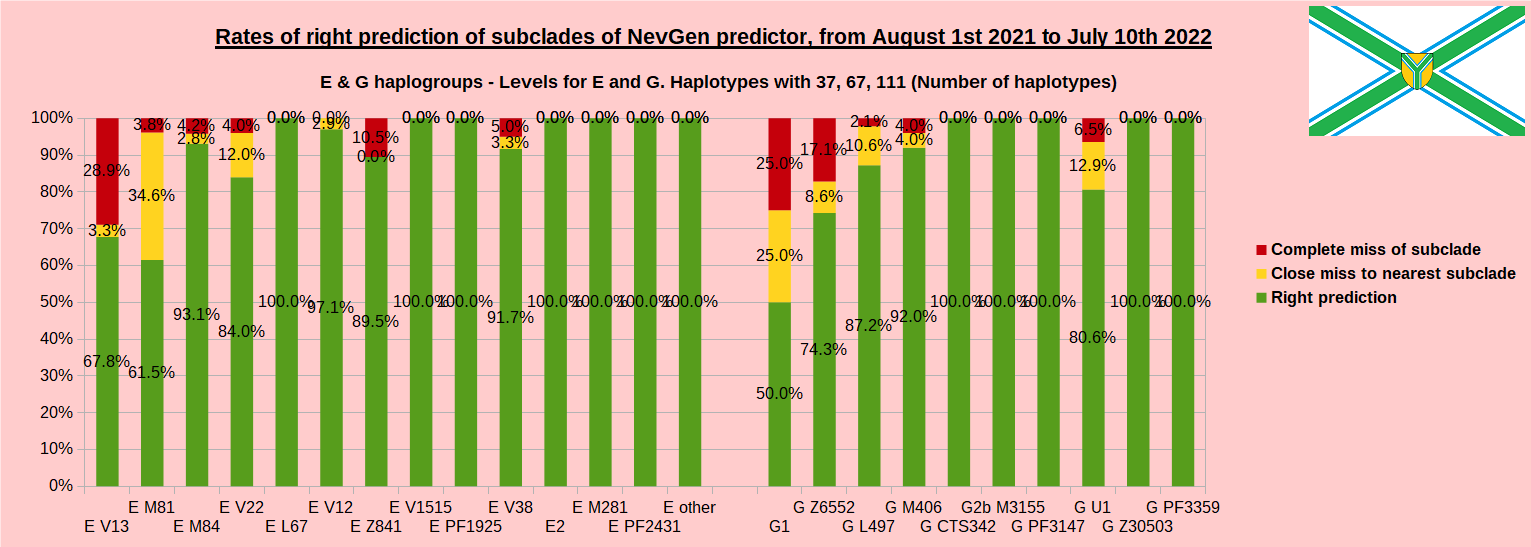
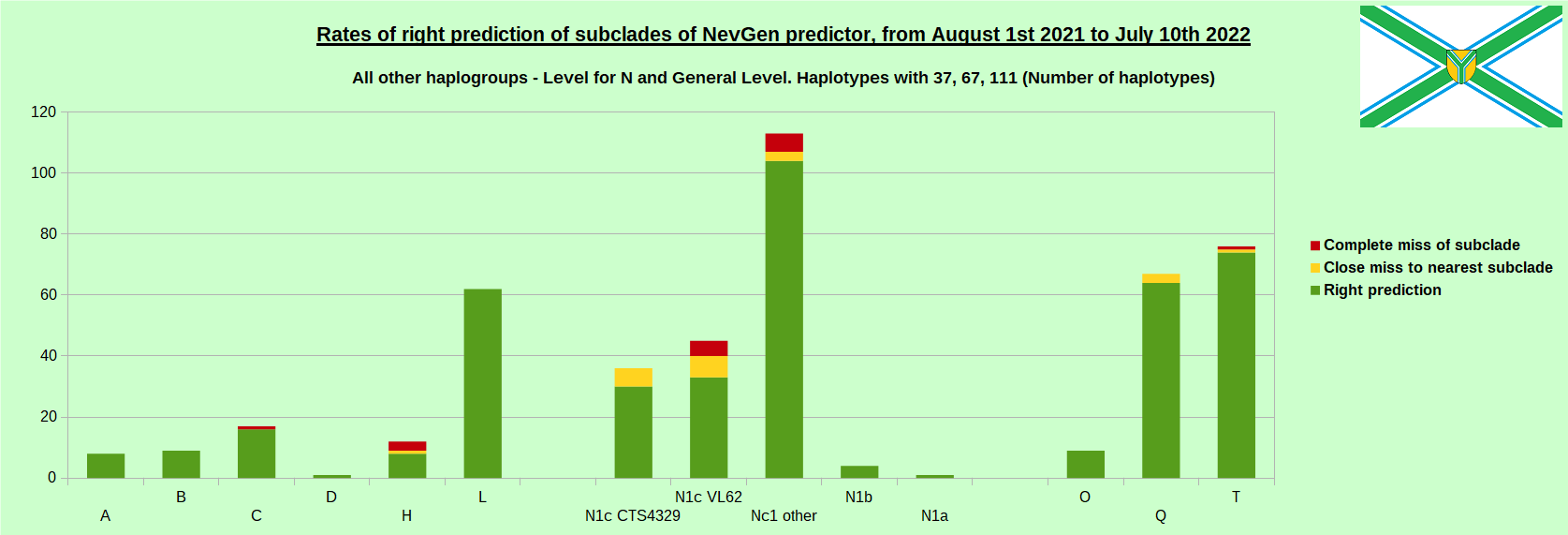
We have prepared the newest results of testing of NevGen Predictor. Time span is from August 1st 2021 to July 10th 2022, little less than year.
As was done in previous years, newly found haplotypes with known deep SNPs are predicted for (their already known) subclade with NevGen Predictor. Statistics of results can be seen on the next eight pictures (four are by number of haplotypes used, and other four by percentages). Haplotypes which do not belong to any of supported subclades (due to small number of available samples) are not used in this statistics. As always, prediction was considered as right if haplotype belongs to subclade which is the first in list (it's already known subclade has greatest probability), and if it's probability is greater than 0 (please note that probability do not need to be 100%, but must be the first and must be bigger than 0).
First is for whole of haplogroup R (R1b, R1a and R2), second is for haplogroups I and J, third is for haplogroups E and G and fourth is for N and all other haplogroups. Percentages of right predictions can be compared to prediction rates of previous year.
Like we said before, to avoid confusion, should be noted that statistics is concerned with prediction of deeper subclades (those that NevGen Predictor supports) of mentioned higher-level haplogroups, not for prediction of higher-level haplogroups themselves. For example, I1’s 88.4% of right subclade predictions is for prediction of subclades of I1, not for prediction of I1 alone, which is easy with at least 25 markers. The same holds for all other haplogroups from pictures, like I2a > L38, I2a > Slavic-Carpathian, I2a > Isles, I2a > M223, J1, E > V13, E > M84, N > VL62, N > CTS4329, R1b > M222, R1a M458, C, L, Q, T, D and so on.
Important thing is that all haplotypes which are less than 9/111, 6/67 or 4/37 close to any haplotype which is part of statistics of it’s already known subclade are excluded from this statistics. That way statistics is more reliable, since such close haplotypes are almost always good predicted. Haplotypes with less than 37 markers were not used in this testing, and majority of them had 111 markers.
Level for R1b-M222 used here is not available in public, since we are not satisfied with it’s results. From statistics for R1b-L21 are excluded all haplotypes of M222 and L226, because they are trivial to predict themselves, even on smaller number of markers (but not their deeper subclades).
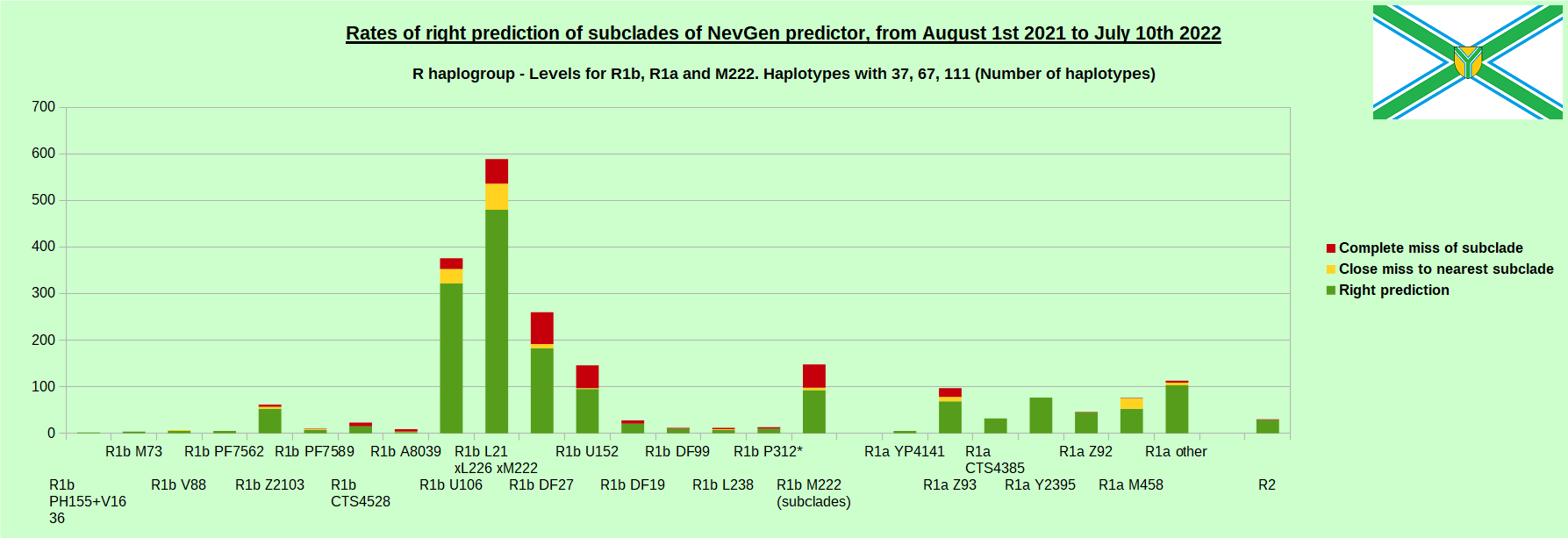
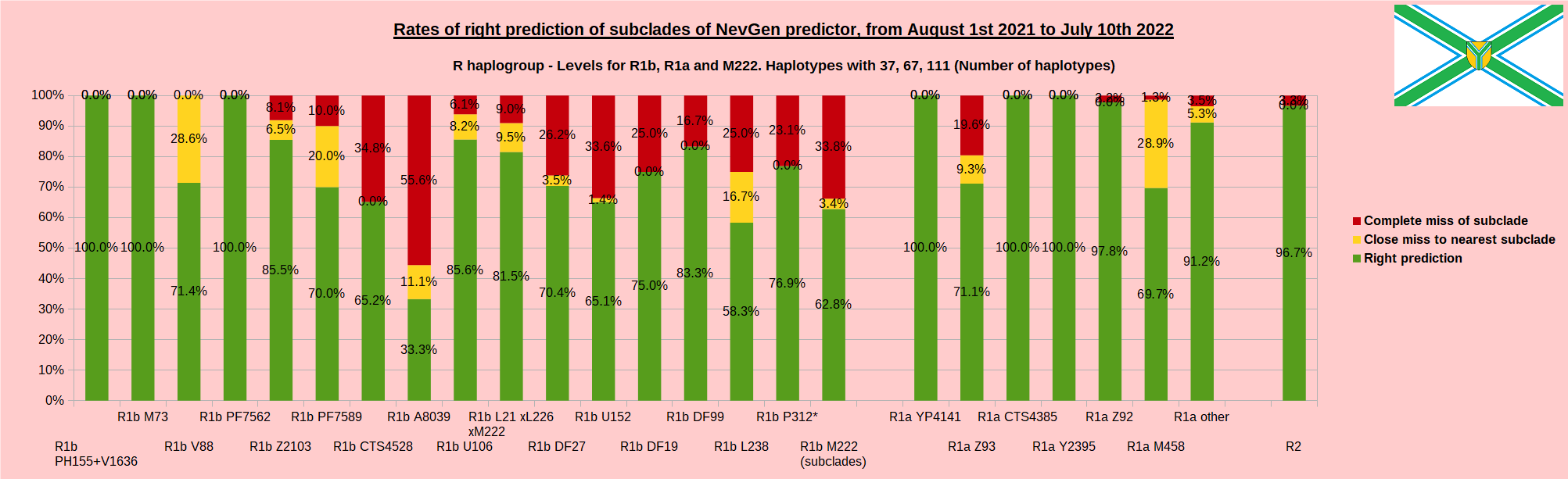
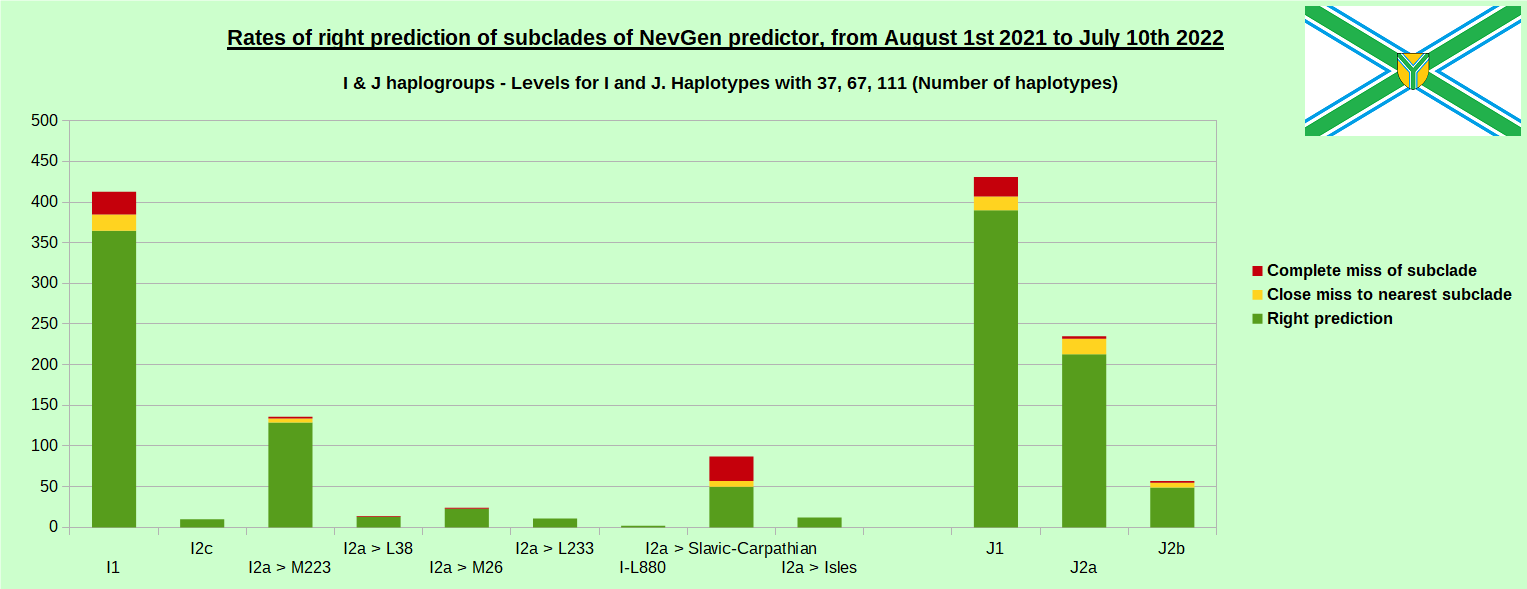
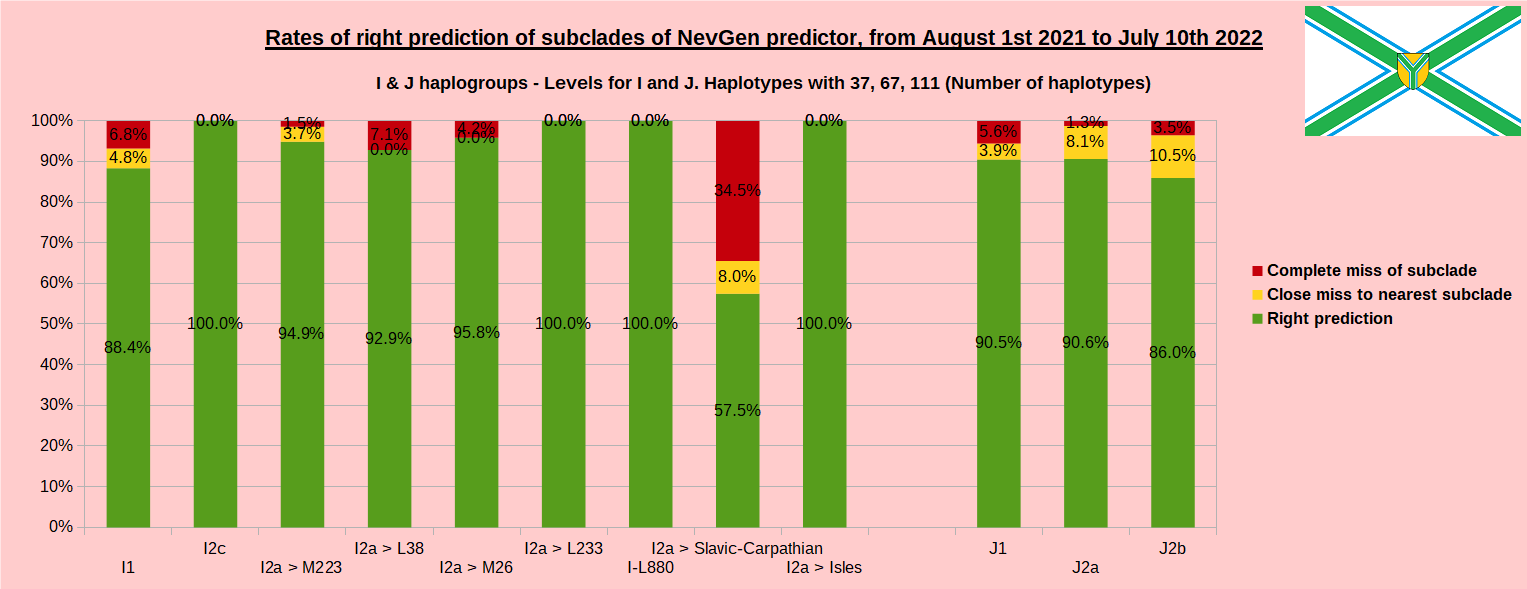
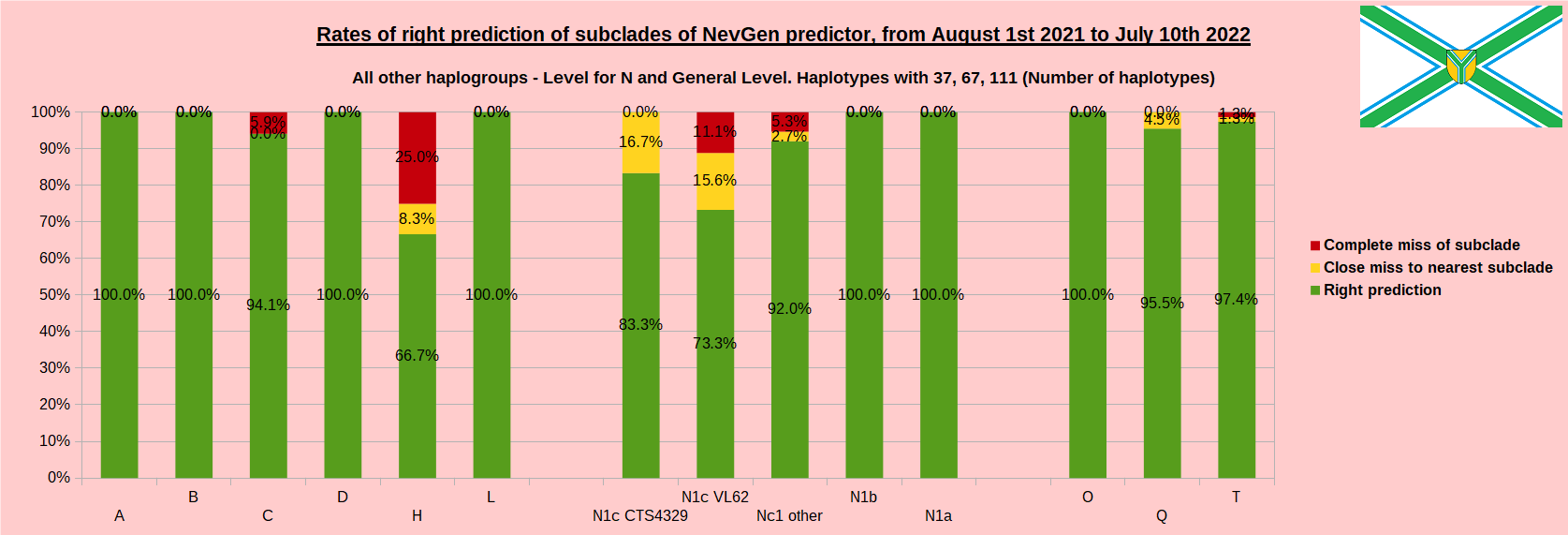
In this year again, the worst prediction rates of major haplogroups are for I2a > Slavic-Carpathian. But this year we have more right predicted than wrong predicted, last year's results for this haplogroup were worse. This year second place as the worst prediction results are again for R1b-U152, 65.1% (two years ago it was the worst), better than last year's 59.5%. I still believe we need many hundreds (if not thousands) of new haplotypes to get satisfying results for U152.
Like in the previous years, with yellow colour we have marked haplotypes which got the most of probability to subclade which is very close, one level below. For example, if our haplotype is by SNP proven to belong to “R1b U106>Z381> Z301>L48> Z9>>Z326>> A5011”, but we got top probability in NevGen for “R1b U106>Z381> Z301>L48> Z9>>Z326>> BY4305”, we than record this testing as “close miss”.
From our experience, subclades under L1335 and FGC11134 (R1b > L21), are not easy to be distinguished, and also Z326 under U106 or L1029 (R1a > M458) or under L258 (I1), and under N-VL62 and N-CTS4329, just to mention some of them.
When comparing with results of previous year, we can see that in many major haplogroups percentages of right-predicted haplotypes increased significantly. Main reason for that is the bug in data organisation which I found and fixed last autumn. In some haplogroups percentages decreased significantly, G-U1 and E-V38, but reason for that is that they were in meantime heavily divided into deeper subclades, which makes prediction harder, so their results of those years are not comparable. Here you can see some of improvements:
- R1a Z93: 51.1% => 71.1%
- R2: 76.2% => 96.7%
- I1: 75.8% => 88.4%
- R1b DF27: 60.5% => 70.4%
- R1b L21: 76.0% => 81.5%
- R1b U152: 59.5% => 65.1%
- R1b U106: 82.4% => 85.6%
- I2a L621 Slavic-Carpathian: 42.3% => 57.5%
- R1b M222: 53.3% => 62.8%
- R1a M458: 58.2% => 69.7%
- I2 M223: 88.8% => 94.9%
- G L497: 76.3% => 87.2%
Out of 228 found STR signatures of length 5, 224 were right (proven by SNP), which gives success rate 98.25%.
Out of 248 found STR signatures of length 4, 232 were right, which gives success rate 93.55%.
Out of 369 found STR signatures of length 3, 323 were right, which gives success rate 87.53%.
Out of 76 found STR signatures of length 1, 70 were right, which gives success rate 92.1%.
Out of 297 found STR signatures of length 2, only 188 were right, which gives success rate of 63.3% This are worst results of all STR signature lengths, I must do something to better filter them, to get more reliable predictions.
Please beware that sum of previous does not need to add to 1008, because for some haplotypes more than one STR signature has been found, and some signatures could not be checked because of missing deeper SNPs.
STR signatures are recognized in all haplogroups and can be found in most of clades, but more often in clades which are older, because their descendants have more diverse and structured haplotype STR values. Here is the list of number of found deeper STR signatures by main haplogroups:
- B: 4/9 = 44.44%
- L: 25/62 = 40.32%
- Q: 21/67 = 31.34%
- J2: 89/292 = 30.48%
- T: 23/76 = 30.26%
- E: 104/363 = 28.65% (V13: 10/90, M84: 20/72, M81: 6/26, V12: 18/34 = 52.94% !!!!, V22: 8/25)
- R2: 7/30 = 23.33%
- R1b: 358/1706 = 20.98% (U106: 81/376, L21: 136/589, M222: 15/148, U152: 28/146, Z2103: 18/62, DF27: 45/260)
- I1: 84/413 = 20.34%
- N: 40/199 = 20.1%
- I2: 59/296 = 19.93% (M223: 37/136, CTS10228 Slavic-Carpathian: just 2/87 = 2.3%!)
- J1: 84/431 = 19.49%
- R1a: 80/446 = 17.94%
- C: 3/17 = 17.65%
- G: 25/164 = 15.24% (L497: 9/47)
Now about long-distance right predictions. That are predictions when haplotypes were right predicted, despite being very far from any sample in our statistics of it’s
subclade. In our new 11-months time-span now we had 344 of them (in last year it was 393), when distance from nearest is at least 30/111. Such cases mostly happened in haplogroups E and J2a, just like in last year.
This year's most distant successful prediction (second best of all times, the best is still 96 markers far) is for 111-markers haplotype of haplogroup E2-M75, which is right predicted despite being 87 markers far from its nearest in training dataset for its haplogroup (it was predicted with only 8.21% probability, but no other clade got more than 0).
Second most distant successful prediction this year is in haplogroup B1 M236, where distance to nearest was 80 (0.92%), and third is in haplogroup E1b1a V38> M329, where distance to nearest was also 80 (but only 0.09% of probability!). Of course, in all cases these were the greatest probabilities given of all clades. In R1b, the greatest distance with right prediction was 54, and haplotype belonged to subclade R1b PH155.
Here you can see the list of such long-distance right predictions, by haplogroup, with distance from nearest and it’s subclade, for those 40 markers and over.
A (5 haplotypes): 57 (A1a M31), 42, 43, 64 (A1b1b2b M13)
B (1 haplotype): 80 (B1 M236)
C (9 haplotypes): 43 (C1 > PH407), 40, 55 (C1a2 V20), 42, 43, 44, 48 (C2 M217> F1067)
D (1 haplotype): 52 (D1b1b CTS6609)
E (70 haplotypes): 47, 61, 72, 73 (E1a M132), 49 (E1b1b M123>M34> M84>> Y14899), 46 (E1b1b M123>M34> Z841>> Y2947), 40, 62 (E1b1b M123>M34> Z841>> Z21466),
45 (E1b1b M84>> PF6751> PF6748 > BY10862), 44, 47 (E1b1b M84>> PF6751> PF6748 >> S1896), 41, 80 (E1b1a V38> M329), 40 (E1b1a V38>> L485), 43, 45 (E1b1a V38>> M4231), 44 (E1b1b V12>CTS693),
40, 43 (E1b1b V12>V32), 57 (E1b1b V1515), 45 (E1b1b V1515> PH1818), 48 (E1b1b V22), 44 (E1b1b V22>> BY7446), 40 (E1b1b V257> PF2431), 75, 87 (E2 M75)
G (18 haplotypes)
H (3 haplotypes)
I1 (4 haplotypes)
I2 (24 haplotypes): 49 (I2a1 L161 ("Isles")> Y13338), 43 (I2a2a M223>L701> L699> S12195), 57 (I2a2a M223>Z161> CTS11871)
J1 (15 haplotypes): 43 (J1a >> P58>> S4924 >> ZS2689), 19/37 (J1a >> PH77), 46 (J1a2 > ZS6599), 62 (J1b F4306>> F1614>> Z2223)
J2 (60 haplotypes): 48 (J2a1 L26>Z500> M92> PF7412), 48 (J2a1 M67>> S25258> SK1336), 47 (J2a1 PF5191>> S15439), 41, 44 (J2a1 Z6046> Y12610), 73 (J2a1 Z6046> Z6048), 40, 43 (J2a1 Z6065> Z7532), 42 (J2a1 Z7671> CTS900> Y11202), 43, 44, 50, 51 (J2a1 Z7700> FGC9883), 40, 46 (J2a1 Z7700> SK1382), 46, 46 (J2a2 PF5008 >> PF5040), 40 (J2a2 PF5008 >> Y29673), 52, 64 (J2a2 PF5008 >> Z39726), 42, 52 (J2b2a M241>Z2444)
L (22 haplotypes): 40, 41, 46 (L1a M27), 40 (L1b M317> M349), 42 (L1b M317> SK1412> SK1414), 43 (L1c M357)
N (1 haplotype)
O (5 haplotypes): 65 (O2a1 F51)
Q (18 haplotypes): 44 (Q L275>> BZ50023), 43, 58, 24/37, 26/37 (Q M346>> M3> M902), 42, 61, 64, 67 (Q M346>> Y4800> F835> L932), 49 (Q M346>> Z780)
R1a (10 haplotypes)
R1b (39 haplotypes): 44 (R1b DF27>Z196> L176.2>CTS4188> S11121), 44 (R1b M73), 54 (R1b PH155), 40 (R1b V1636), 40, 45 (R1b V88 >> V1589)
R2 (19 haplotypes): 41, 41, 43, 47, 51, 54, 54 (R2 M479), 40, 42 (R2 M479 >> FGC13203)
T (20 haplotypes): 41, 46, 19/37 (T >> CTS11451> PF7455), 40, 40 (T >> CTS11451> Y4119), 46 (T >> CTS11451> Y4119> Y16244), 43 (T >> CTS11451> Y4119>> Y9109)